Retina DS Cell Fisher Information Codes
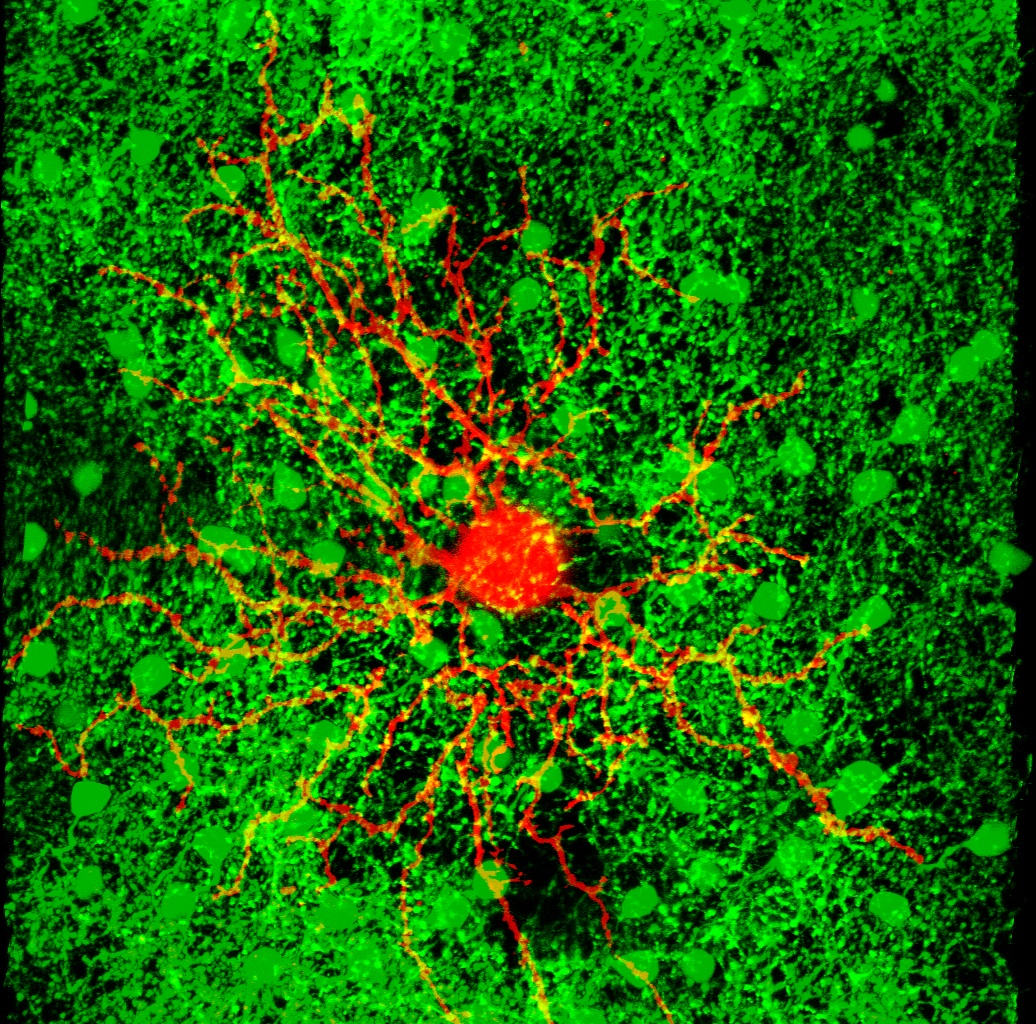 This page contains the computer codes to run a computational model of the direction selective ganglion cells (DS cells) of the mouse retina, like the orange dye-filled cell in the picture. It also contains the experimental data upon which that model is based. (Links to download the codes and data are at the bottom of this page.)
This page contains the computer codes to run a computational model of the direction selective ganglion cells (DS cells) of the mouse retina, like the orange dye-filled cell in the picture. It also contains the experimental data upon which that model is based. (Links to download the codes and data are at the bottom of this page.)
Along with simulating the DS cells’ responses to moving stimuli, there are codes to compute the amount of information that these cells convey to the brain about the stimulus. The codes compute the information values for both the “normal” responses of the model — which closely match our experimental observations from the mouse retina — and for models in which the correlations between cells are either removed, or are manipulated so as not to depend on the stimulus.
The model, and the experiments, are described in more detail in our 2016 Neuron paper Direction-selective circuits shape noise to ensure a precise population code.
At the bottom of this page, you will find .zip files containing everything you need to run the DS population model, and to reproduce the theoretical Fisher information calculations, in addition to the spiking responses recorded simultaneously from pairs of DS cells. Below is a description of each file, the hierarchy of files, etc. Please cite our Neuron paper for any work derived from use of this code and/or data. All materials are provided as-is, with no guarantees. Please alert me to any issues / bugs that you encounter Also, if you have any questions, don't hesitate to ask.
Here is the code to run the DS cell model fitted to the experimental data, and to compute the amount of information the modeled population conveys about the stimulus.File details:
- The script ooDS_model.m loads up the model parameters (ooDS_params.mat) and runs the model, returning the activity statistics for different stimuli. - ooDS_Fisher.m calls ooDS_model.m, and then uses the activity stats to compute the Fisher info. Results should be able to reproduce Fig. 6E. To Run the analysis in matlab: >> ooDS_FisherHere is the code to perform the theoretical calcuations about how stimulus-dependent correlations between neurons affect information transmission from the eyes to the brain.
File details:
- The script TC.m contains the tuning curve shapes. - TC.m is called by do_FI_calc_stimdep.m, which generates responses from the tuning curves, and computes the covariance matrices using Eq. 2 of the paper (assuming Poisson noise). It then computes the Fisher info for the case of stim dependent correlations, and for the case of “matched” constant correlations. - There are two "looper" scripts that repeat the calculation for different population sizes and correlation strengths. looper_FI_HOMOG.m specifies homogeneous tuning curves, whereas looper_FI_HETEROG.m specifies heterogeneous ones. To Run for heterogeneous tuning curves (note, this may take several minutes to run): >> looper_FI_HETEROG.m To Run for homogeneous tuning curves (note, this may take several minutes to run): >> looper_FI_HOMOG.mHere are the extracellular spiking recordings used to generate Fig. 1 of the paper, and to fit the model in Fig. 6
File details:
- readme.rtf contains a description of the matlab files and their formatting -DSpairs_spikeData.mat contains the data